sciRNA-seq decoding
This method was used to generate a transcriptome map of C. elegans L2 larvae at the single-cell level. Cells or nuclei are isolated from whole organisms, tissues, or cell culture. Individual cells/nuclei are pooled and distributed to 96- or 384-well dishes at around 10-100 cells per well. mRNAs are reverse transcribed in Plate 1 using barcoded primers (96 RT barcodes, one per well). Each barcoded primer also carries a unique molecular index (UMI). Cells are then repooled and split again into new plates and second-strand synthesis, tagmentation, and PCR are then performed. Plate 2 adds the standard i5 and i7 dual indexed (8 x 12) barcodes. The method is described in Comprehensive single-cell transcriptional profiling of a multicellular organism Cao et al. Science 2017, 357:661-667 | DOI: 10.1126/science.aam8940
If the results are written to SAM, BAM or CRAM the raw uncorrected cellular barcode sequence is written to the CR auxiliary tag, the corresponding quality scores are written to the CY auxiliary tag, the corrected cellular barcode sequence is written to the CB auxiliary tag and the probability that the Cellular barcode decoding is incorrect is written to the XC auxiliary tag.
The uncorrected mollecular barcode (UMI) sequence is written to the RX auxiliary tag and the corresponding quality scores are written to the QX auxiliary tag.
Read Anatomy
The final PCR products submitted for sequencing are composed as follows:
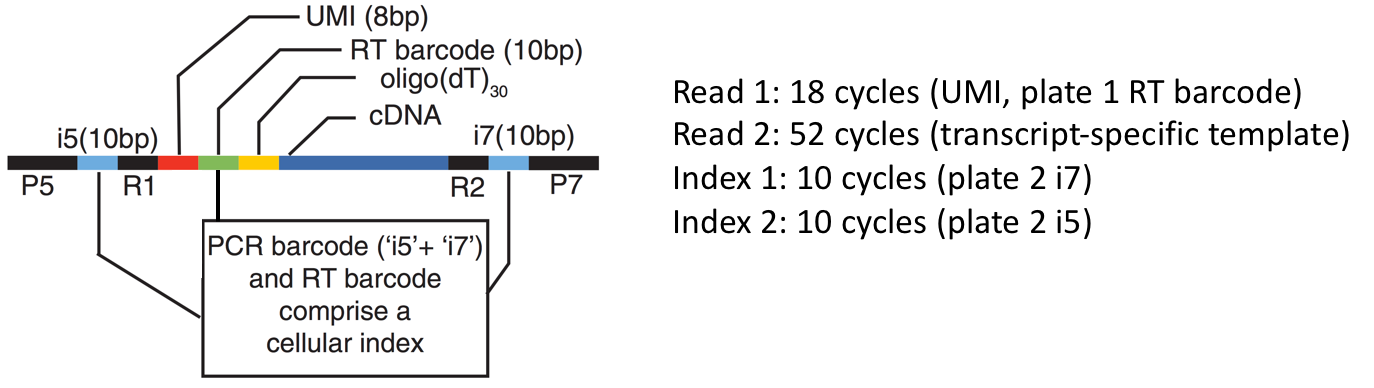
Input Read Layout
Base calling with bcl2fastq will produce 4 files per lane:
HGGKLBGX2_L001_R1_001.fastq.gz: Read 1, starting from the beginning of the insert fragment (“top” strand).HGGKLBGX2_L001_I1_001.fastq.gz: Index 1, the i7 index.HGGKLBGX2_L001_I2_001.fastq.gz: Index 2, the i5 index.HGGKLBGX2_L001_R2_001.fastq.gz: Read 2, starting from the other end of the insert fragment (“bottom” strand); note that this sequence is reverse complemented relative to the Read 1 sequence.
"input": [ "HGGKLBGX2_L001_R1_001.fastq.gz", "HGGKLBGX2_L001_I1_001.fastq.gz", "HGGKLBGX2_L001_I2_001.fastq.gz", "HGGKLBGX2_L001_R2_001.fastq.gz" ]declaring input read segments 2 biological and 2 technical sequences are often found in 4 FASTQ files produced by bcl2fastq base calling.
Read segments emerge from the sequencer in the same order as a simple dual-indexed run (R1, I1, I2, R2). We wish to extract the UMI (8bp) and RT barcode (10bp) from Read 1, the cDNA template sequence from Read 2 (50 bases), the i5 index (10bp) from the Index 2 read, and the i7 index (10bp) from the Index 1 read.
Token expression Segment index First Last Length Description 0::80078UMI on segment in R1 0:8:18081710RT barcode on segment in R1 1::1010910i7 barcode on segment in I1 2::1020910i5 barcode on segment in I2 3::50304950template on segment in R2 Tokenization patterns for decoding tha various artifacts of a sciRNA-seq protocol.
{ "cellular": [ { "algorithm": "pamld", "base": "rt_c_elegans", "comment": "Plate 1 RT barcode", "confidence threshold": 0.99, "noise": 0.05, "transform": { "token": [ "0:8:18" ] } }, { "algorithm": "pamld", "base": "nextera_dual_barcode_set", "comment": "Plate 2 PCR barcode (i5 + i7)", "confidence threshold": 0.99, "noise": 0.05, "transform": { "token": [ "1:0:10", "2:0:10" ] } } ], "import": [ "HGGKLBGX2_core.json" ], "input": [ "HGGKLBGX2_L001_R1_001.fastq.gz", "HGGKLBGX2_L001_I1_001.fastq.gz", "HGGKLBGX2_L001_I2_001.fastq.gz", "HGGKLBGX2_L001_R2_001.fastq.gz" ], "molecular": [ { "comment": "The UMI is in the first 8 bases of Read 1", "transform": { "token": [ "0::8" ] } } ], "template": { "transform": { "token": [ "3::50" ] } } }decoding both combinatorial barcodes in one run HGGKLBGX2_l01_cellular.json will decode both the RT and nextera barcodes in one run and populate the cellular and molecular SAM auxiliary tags. It will emit a single segment with the first 50 nucleotides on the forth segment in R2. The list of possible values for the RT and nextera barcodes are defined in the imported HGGKLBGX2_core.json configuration file and are used in HGGKLBGX2_l01_cellular.json by inheritence using
"base": "rt_c_elegans"and"base": "nextera_dual_barcode_set".